Junghyun has been selected as a scholarship recipient by the JW이종호재단!
Congratulations to Junghyun for being awarded the Basic Research Scientist Residential Support from the JW Foundation in 2023!! We love you~

Soyeon has been accepted with a full scholarship into the Ph.D. program at the University of Cambridge, affiliated with Wellcome Sanger Institute!!
This is indeed great news, taking into account the immense efforts she has put in.
We hope the days ahead of her new start will be filled with happiness.

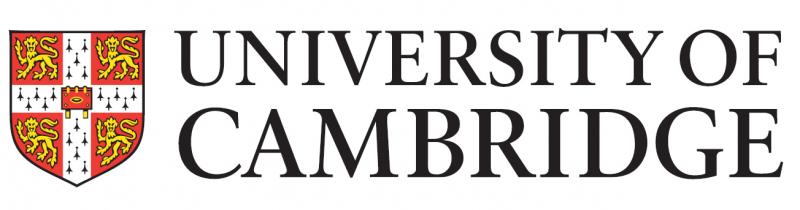
Prof.Siyuan Zheng (from U of Texas, San Antonio) visited Our Lab!!
Thank you for visiting our lab
Prof.Roel Verhaak visited Our Lab!!
Thank you for visiting our lab


Prof.Kim has been invited to the 43rd Frontier Scientists Workshop (“Future Trends in Genomic Mutation and Disease Research”)
Future Trends in Genomic Mutation and Disease Research which will be held in San Diego, USA by the Korean Academy of Science and Technology (KAST)

"2022년도 SKKU-SMC 미래융합연구 과제 선정"
Our lab has been selected for 2022년도 SKKU-SMC 미래융합연구

Happy Birthday, Jiwon!!
Happy birthday Jiwon!! Our lab's lovely quokka girl 😘 🥰 !! We love you~

"2022년도 STEAM연구사업 미래유망융합기술파이오니어 과제 선정"
Our lab has been selected for 2022년도 STEAM연구사업 (미래유망융합기술파이오니어 : 과학난제 도전형)


Soyeon’s visiting JAX!!!
We hope you will learn well and come back!! Have a safe flight and all the best to your new journey!! We love you~<3
We celebrated Jaeho's birthday!!
Happy birthday Jaeho!! We are so happy that you are one of lab members!! We love you~<3

We celebrated Junyong's birthday!!
Happy birthday Jun Yong!! Our lab's very cute shy boy 😇 ~!! We love you~<3

We celebrated YoonJoo's birthday!!
Happy birthday Yoon Joo~!! Our lab's all-rounder 😝 !! We love you~<3

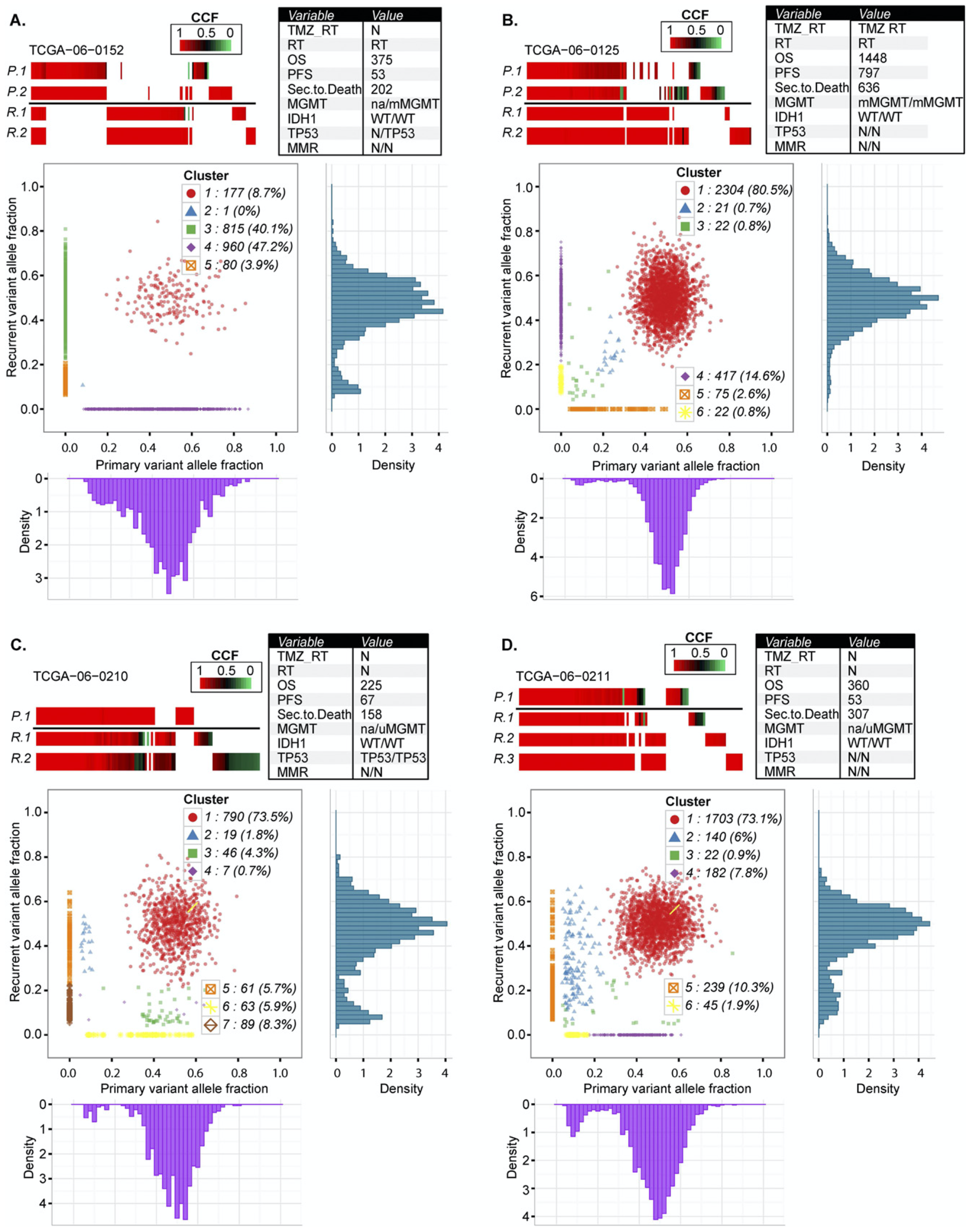
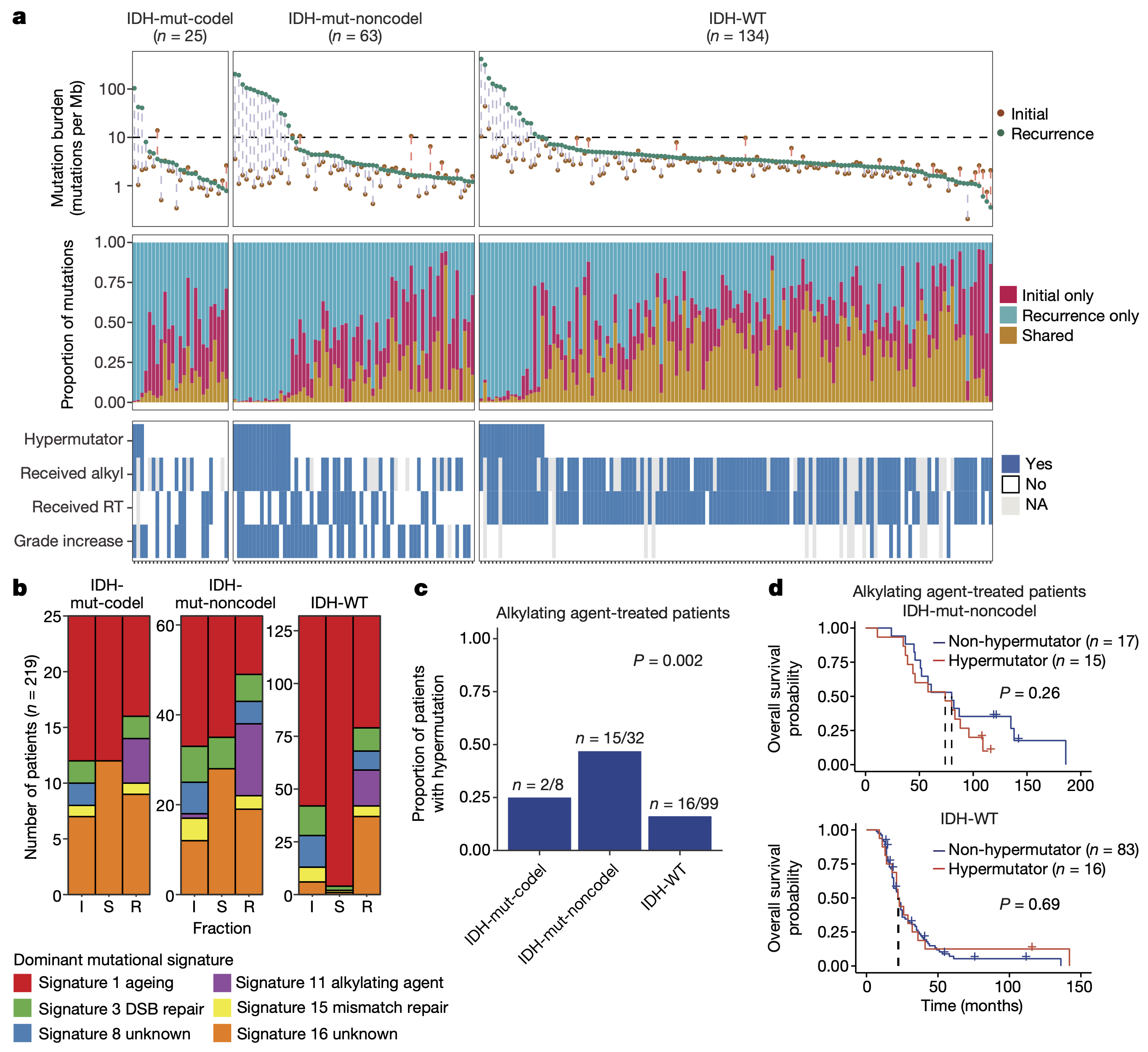
"Glioma progression is shaped by genetic evolution and microenvironment interactions"
Varn et al. Integrating longitudinal transcriptomic and genomic data from paired diffuse glioma samples with complementary single-cell RNA-seq and multiplex immunofluorescence datasets reveals recurrence-associated genetic and microenvironmental changes that are dependent on IDH mutation status.
Verhaak, et al. Cell 9 June 2022. Full text article
We celebrated Harold's birthday!!
Happy birthday Harold~!! Our lab's king king 🤴 !!! Please continue to be Our leader~~!! We love you~<3

We celebrated 2nd Teacher's Day in our Lab !!
Happy Teacher’s Day, we had 2nd teacher’s day celebration with Prof. Kim.


Soyeon was awarded BVIC !!!
Congratulations, Soyeon, for awarded the “SKKU BIO-GLOBAL VALUE INNOVATIVE CREATOR (BVIC)” !!!

Junghyun was awarded Graduate Scholarship !!!
Congratulations, JungHyun, for awarded the “Kwanjeong Educational Foundation” !!!
We celebrated Eunchae's birthday!!
Happy birthday Eunchae~!! Our lab's Pokemon!!! ~ We are so happy that you are one of lab members!! We love you~<3
We celebrated Dongjoo's birthday!!
Happy birthday Dongjoo~!! We are so happy that you are one of lab members!! We love you~<3

We celebrated Yoonjoo,Junghyun's college graduation!!
Congratulations on your graduation, We hope you'll be happy in the future! Our lab's 99s!! We love you guys~<3


Happy Birthday, Steve
Happy birthday Steve!! We hope you go back to 18 Steve~ Once again, happy birthday to our singularity&docker expert! We love you<3

We celebrated Hyunjeong's birthday!!
Happy birthday hyunjeong, our lab's squarrel! We are so happy that you are one of lab members!! We love you~<3
We celebrated Daeho's birthday!!
Happy birthday Daeho!! We are so happy that you are one of lab members!! We love you~<3

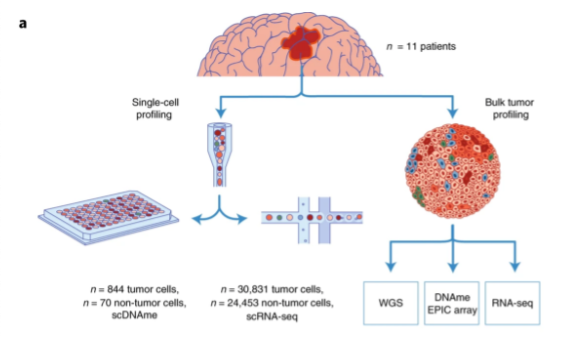
Single-cell multimodal glioma analyses identify epigenetic regulators of cellular plasticity and environmental stress response
Glioma intratumoral heterogeneity enables adaptation to challenging microenvironments and contributes to therapeutic resistance. We integrated 914 single-cell DNA methylomes, 55,284 single-cell transcriptomes and bulk multi-omic profiles across 11 adult IDH mutant or IDH wild-type gliomas to delineate sources of intratumoral heterogeneity. We showed that local DNA methylation disorder is associated with cell–cell DNA methylation differences, is elevated in more aggressive tumors, links with transcriptional disruption and is altered during the environmental stress response. Glioma cells under in vitro hypoxic and irradiation stress increased local DNA methylation disorder and shifted cell states. We identified a positive association between genetic and epigenetic instability that was supported in bulk longitudinally collected DNA methylation data. Increased DNA methylation disorder associated with accelerated disease progression and recurrently selected DNA methylation changes were enriched for environmental stress response pathways. Our work identified an epigenetically facilitated adaptive stress response process and highlights the importance of epigenetic heterogeneity in shaping therapeutic outcomes.
Kevin C. Johnson, Kevin J. Anderson, Hoon Kim, Paul Robson & Roel G. W. Verhaak, et al. nature genetics 30 September 2021. Full text article
We celebrated Youngjin's wedding!!
Congratulations on your wedding, Youngjin! We hope you have a happy day~

We celebrated Junyong's Birthday in our lab!!
Happy birthday Junyong!! he is our 1st undergraduate intern!! We are so happy that you are one of lab members!! We love you <3.

We celebrated Heesuk's BirthDay during KSBMB international Conference!!
Happy birthday Heesuk!! He is our 1st member of Computational Biomedicine Lab. We are so happy that you are one of lab members!! We love you <3.

We celebrated 1st Teacher's Day in our Lab !!
Happy Teacher’s Day, we had 1st teacher’s day celebration with Prof. Kim.
Soyeon was awarded Graduate Scholarship !!!
Congratulations, Soyeon, for awarded the “Graduate scholarship student in the second semester of 2021” !!!

Jun-yong was awarded National Scholarship !!!
Congratulations, Jun-yong, for awarded the prestigious “National Academic Excellence Scholarship (Science and Engineering)” !!!

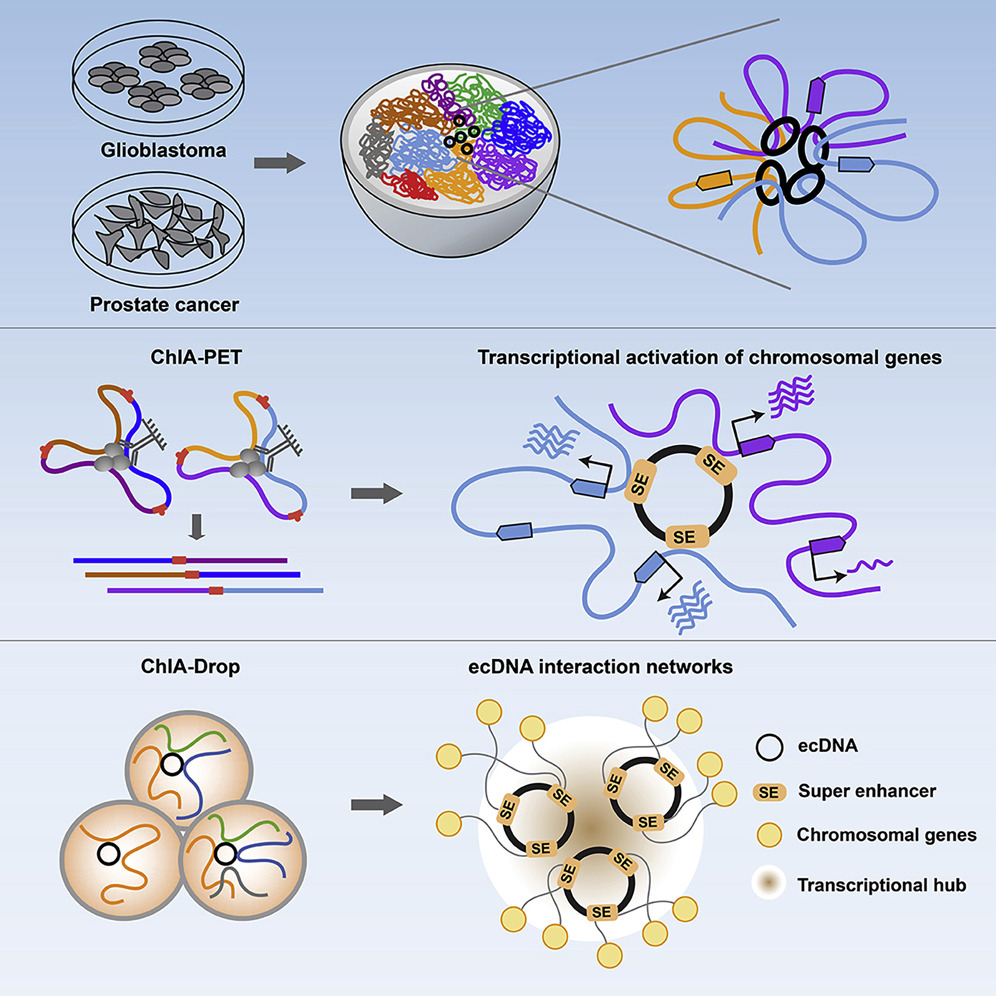
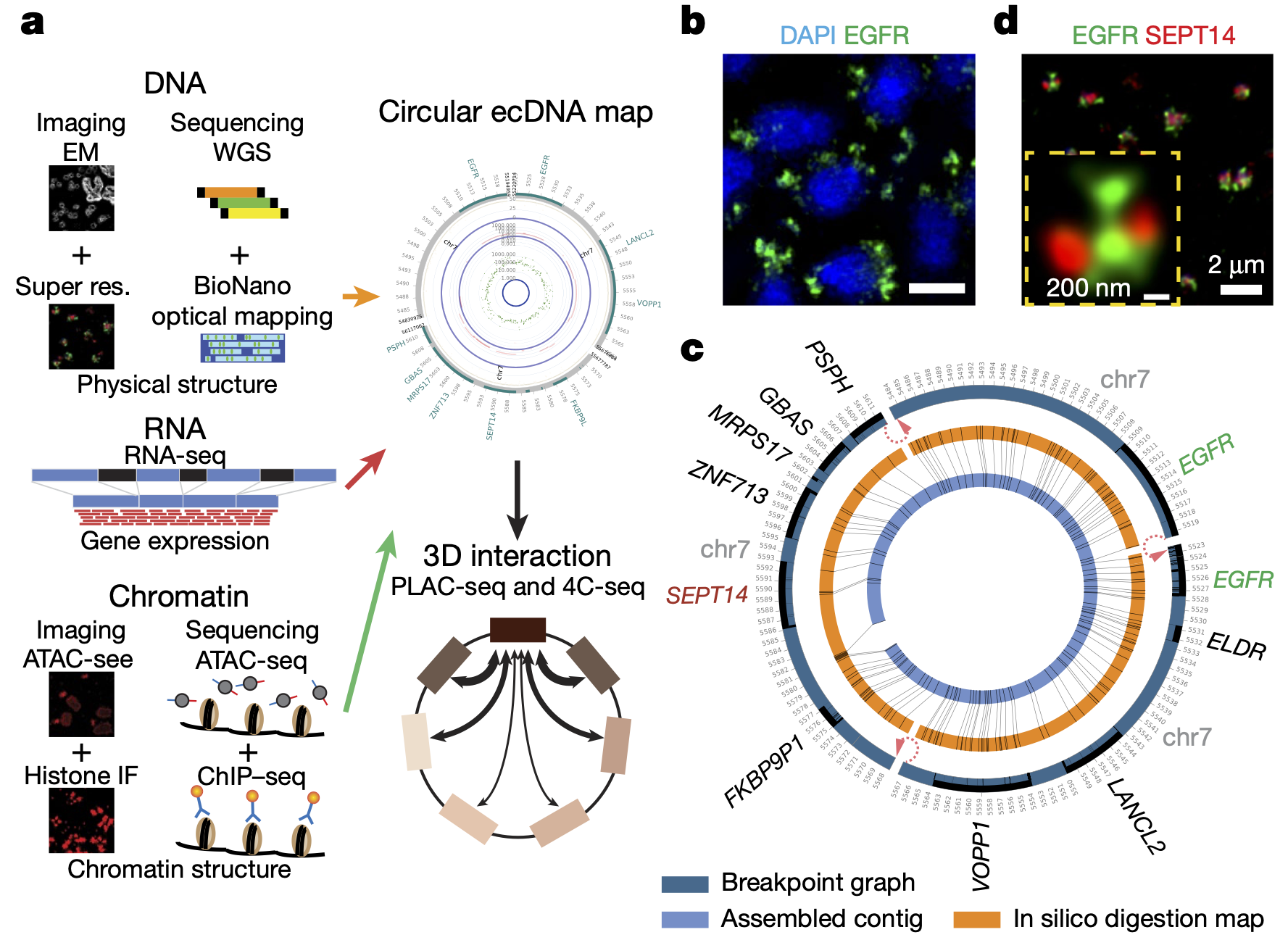
Extrachromosomal DNA functions as mobile enhancers !!!
Congratulations, Yanfen & Amit !!! Zhu et al. report the chromatin connectivity networks of circular and extrachromosomal DNA elements (ecDNA) in cancer, revealing that ecDNAs can function as mobile super-enhancers, which drives genome-wide transcriptional amplification, including that of oncogenes. These findings support an expanded role for ecDNA in trans-regulating chromosomal genes in promoting tumor growth.
NIH/NCI R21 grant application score was posted (top 3 %)
Hoon’s NIH/NCI R21 grant application about extrachromosomal DNA received the top 3 % with impact score 24. It is the most likely to be within the funding line !!!

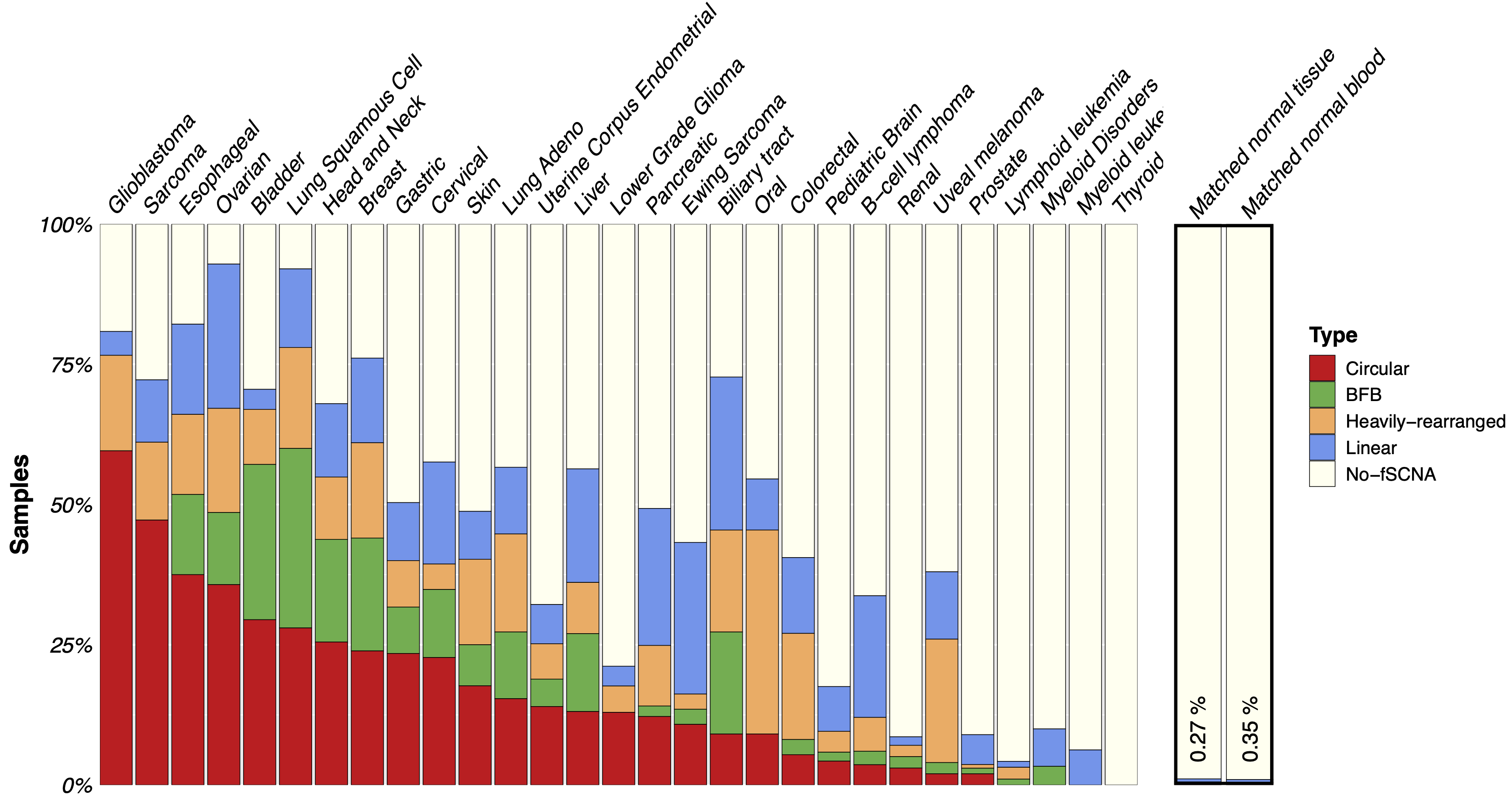
Pan-Cancer analysis of extrachromosomal DNA amplification
Follow up to our previous work on extra-chrmosomal DNA (ecDNA) amplifications in glioblastoma, Hoon Kim led the pan-cancer analysis of ecDNA amplifications in 3,212 tumor and 1,810 normal samples across 29 cancer types. Read more about our work at Nature Genetics and in the press release.
Kim, H., Nguyen, N. P., Turner, K., Wu, S., Gujar, A. D., Luebeck, J., Liu, J., Deshpande, V., Rajkumar, U., Namburi, S., Amin, S. B., Yi, E., Menghi, F., Schulte, J. H., Henssen, A. G., Chang, H. Y., Beck, C. R., Mischel, P. S., Bafna, V., & Verhaak, R. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nature Genetics 08/2020. https://pubmed.ncbi.nlm.nih.gov/32807987/
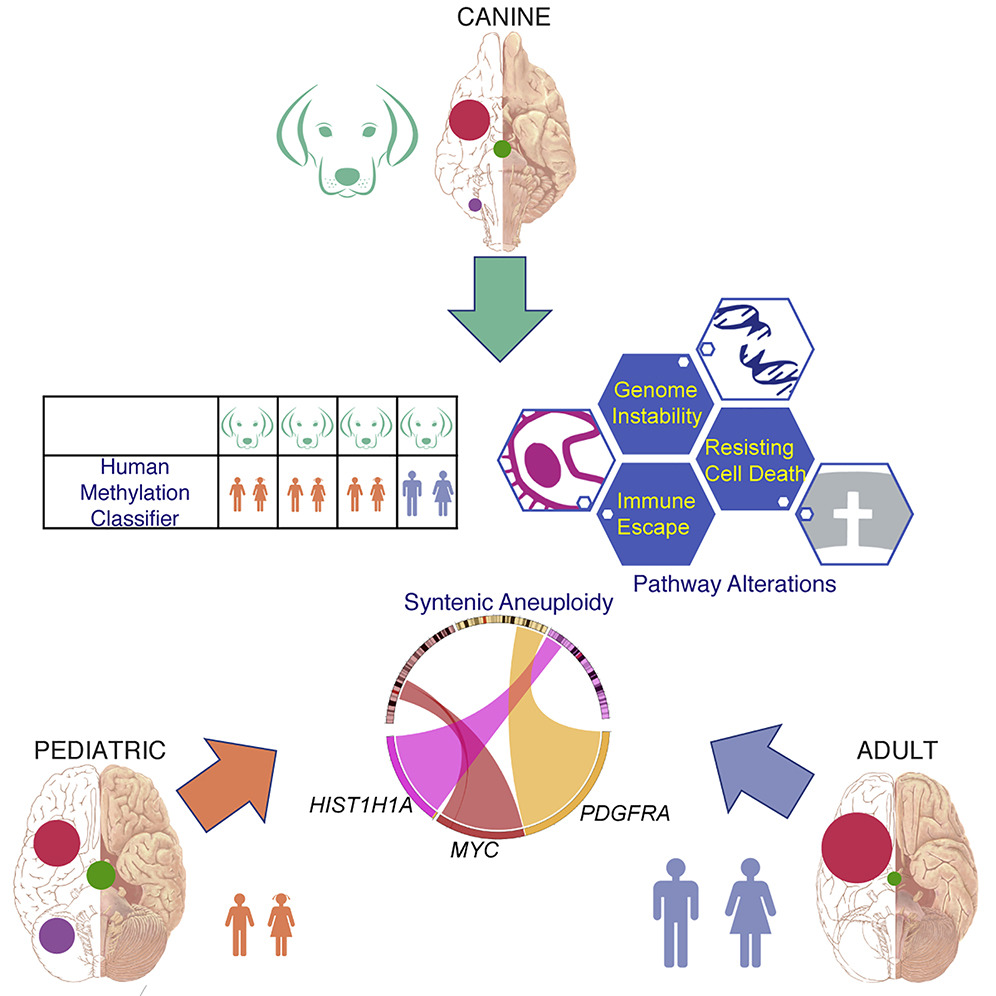
Learning from spontaneous gliomas in companion dogs
Our paper focusing on comparative oncology of cross-species gliomas is out today in Cancer Cell. Dogs get spontaneous tumors similar to humans in an immuno-competent environment. We sought to trace molecular life history of gliomas across two species, i.e., to identify common set of somatic alterations and underlying evolutionary conserved mutational processes. We also queried age-related impact on development of glioma in canines vs. human pediatric and adult patients of glioma to show if canine gliomas resemble more of human pediatric or adult gliomas.
We performed genomic, epigenomic, and transcriptomic characterization of sporadic glioma in dogs (n=83), and compared those to available genomic profiling of patients of human pediatric and adult gliomas. Canine gliomas were marked by significantly lower somatic mutation burden than human adult gliomas but similar to human pediatric gliomas. Despite lower somatic mutation burden, gliomas across canine and human pediatric and adult patients were marked by presence of recurrent, hot-spot, somatic mutations in known glioma drivers like PDGFRA, PIK3CA, and IDH1, suggesting convergent evolution of cross-species gliomas. Overall, gliomas in dogs, despite their prevalence in adult dogs, resembled human pediatric gliomas by mutational burden and DNA methylation.
Comparative oncology of spontaneous cancer in companion dogs can yield not only insights into evolutionary origins of cancer but also provide strong rationale to conduct parallel clinical trials in immuno-competent companion dogs for potential benefits to both, humans and their best friend!
For more, please read our paper here. We also provide companion website, https://canineglioma.verhaaklab.com, that hosts extended methods and code to generate figures.
Samirkumar B. Amin, Kevin J. Anderson, C. Elizabeth Boudreau, Emmanuel Martinez-Ledesma, Emre Kocakavuk, Kevin C. Johnson, Floris P. Barthel, Frederick S. Varn, Cynthia Kassab, Xiaoyang Ling, Hoon Kim, Mary Barter, Ching C. Lau, Chew Yee Ngan, Margaret Chapman, Jennifer W. Koehler, James P. Long, Andrew D. Miller, C. Ryan Miller, Brian F. Porter, Daniel R. Rissi, Christina Mazcko, Amy K. LeBlanc, Peter J. Dickinson, Rebecca A. Packer, Amanda R. Taylor, John H. Rossmeisl Jr, Kevin D. Woolard, Amy B. Heimberger, Jonathan M. Levine, Roel G. W. Verhaak.
Comparative Molecular Life History of Spontaneous Canine and Human Gliomas.
Cancer Cell 2020;37:243–57.e7. doi: 10.1016/j.ccell.2020.01.004.
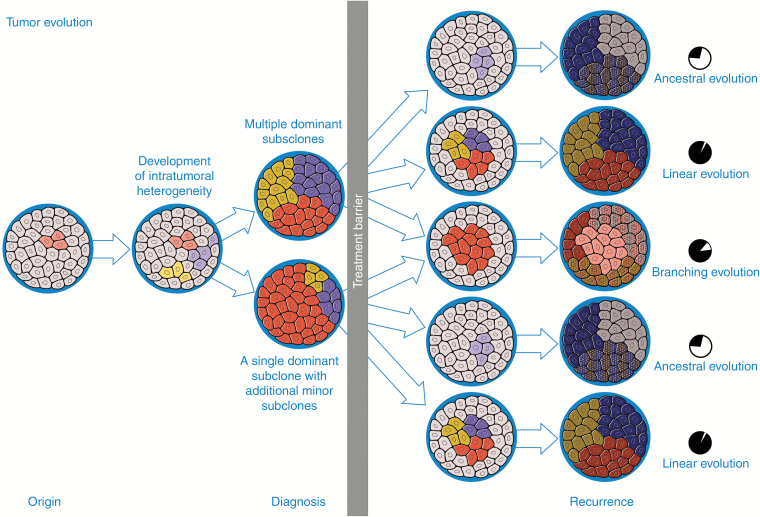
Glioma evolution over time characterized
Floris Barthel, Kevin Johnson are the lead authors on a new study on longitudinal analysis of glioma in adult patients, with contributions from many lab members. This is a first seminal study that leverages the GLASS Data Resource and the first publication in collaboration with the GLASS Consortium. We identified multiple types of evolution and/or treatment associated changes in the glioma genome over time. This seminal work was published in Nature.
Barthel FP, Johnson KC, Varn FS, Moskalik AD, Tanner G, Kocakavuk E, Anderson KJ, Abiola O, Aldape K, Alfaro KD, Alpar D, Amin SB, Ashley DM, Bandopadhayay P, Barnholtz-Sloan JS, Beroukhim R, Bock C, Brastianos PK, Brat DJ, Brodbelt AR, Bruns AF, Bulsara KR, Chakrabarty A, Chakravarti A, Chuang JH, Claus EB, Cochran EJ, Connelly J, Costello JF, Finocchiaro G, Fletcher MN, French PJ, Gan HK, Gilbert MR, Gould PV, Grimmer MR, Iavarone A, Ismail A, Jenkinson MD, Khasraw M, Kim H, Kouwenhoven MCM, LaViolette PS, Li M, Lichter P, Ligon KL, Lowman AK, Malta TM, Mazor T, McDonald KL, Molinaro AM, Nam DH, Nayyar N, Ng HK, Ngan CY, Niclou SP, Niers JM, Noushmehr H, Noorbakhsh J, Ormond DR, Park CK, Poisson LM, Rabadan R, Radlwimmer B, Rao G, Reifenberger G, Sa JK, Schuster M, Shaw BL, Short SC, Smitt PAS, Sloan AE, Smits M, Suzuki H, Tabatabai G, Van Meir EG, Watts C, Weller M, Wesseling P, Westerman BA, Widhalm G, Woehrer A, Yung WKA, Zadeh G, Huse JT, De Groot JF, Stead LF, Verhaak RGW; GLASS Consortium.
Longitudinal molecular trajectories of diffuse glioma in adults.
Nature. 2019 Dec;576(7785):112-120. doi: 10.1038/s41586-019-1775-1.
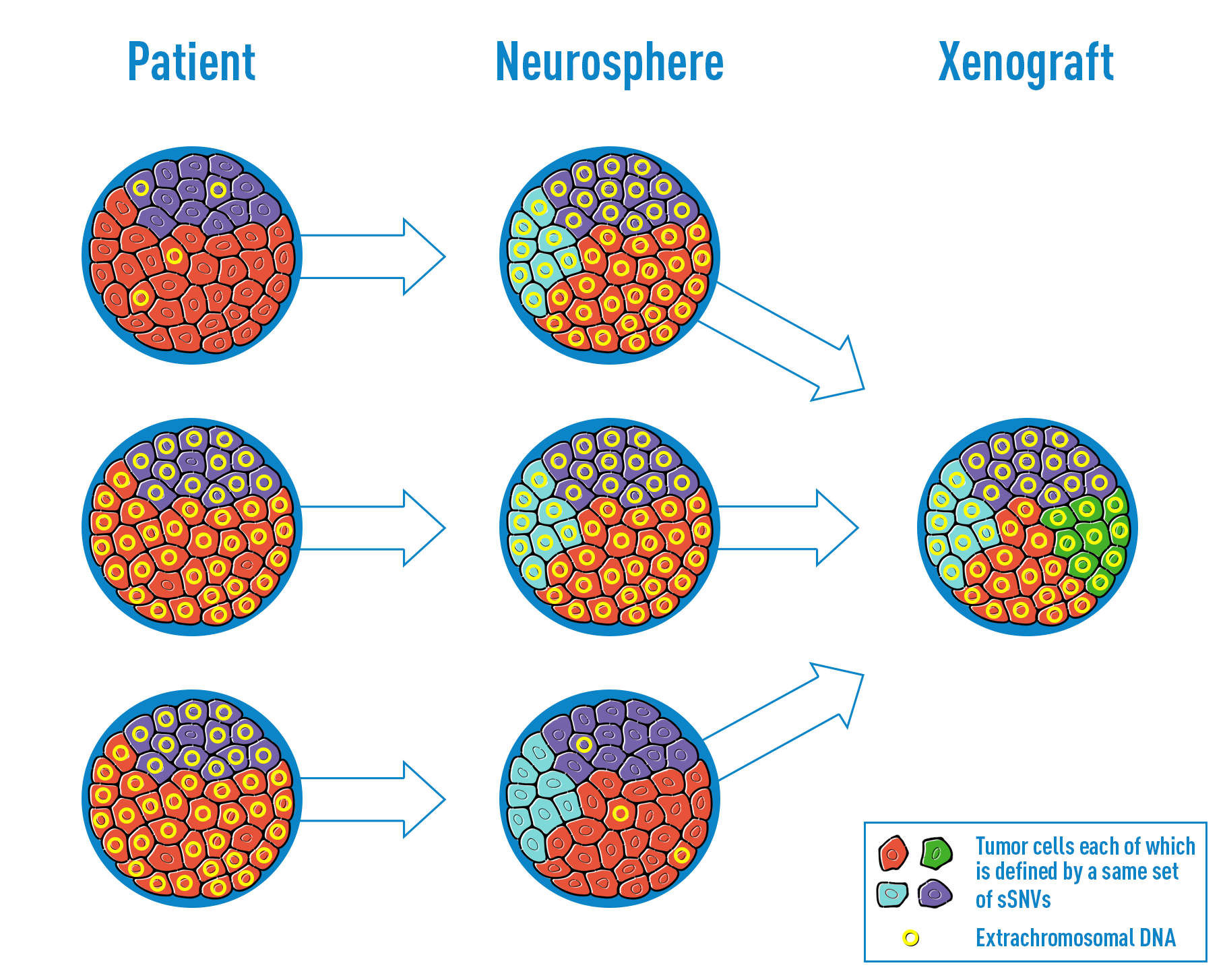
Circular ecDNA promotes accessible chromatin and high oncogene expression
deCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet 2018. PMID: 29686388
GLASS Data Resource released
The mission of the Glioma Longitudinal Analysis (GLASS) Consortium is to create a longitudinal molecular reference data set that can be used to characterize and understand the processes that drive glioma evolution and therapy resistance. Towards that goal, the first GLASS Data Resource was released today, consisting of multiple time-point exome and/or whole genome sequencing profiles from 257 glioma patients.
Patient data and matching clinical annotations were provided by many labs across the world; data pre-processing was all done by the Verhaak laboratory. For more information, see www.glass-consortium.org.

Announcing Glioma Longitudinal AnalySiS (GLASS) Consortium
The Glioma Longitudinal AnalySiS (GLASS) Consortium announces itself to the world in Neuro-Oncology, with Roel as corresponding author.
GLASS Consortium. Glioma Through the Looking GLASS: Molecular Evolution of Diffuse Gliomas and the Glioma Longitudinal AnalySiS Consortium. Neuro Oncol 2018. PMID: 29432615
extrachromosomal DNA amplification in GBM
A formative publication for the Verhaak lab, our first publication fully focused on extrachromosomal DNA amplification, with co-first author Hoon, proudly published in Nature Genetics.
deCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet 2018. PMID: 29686388
Next chapter of the Expression Subtypes of Glioblastoma
Qianghu build the next chapter of the “expression subtypes of glioblastoma” saga, building on Roel’s GBM subtypes postdoc paper and appropriately published in Cancer Cell.
Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017;32:42–56.e6. PMID: 28697342

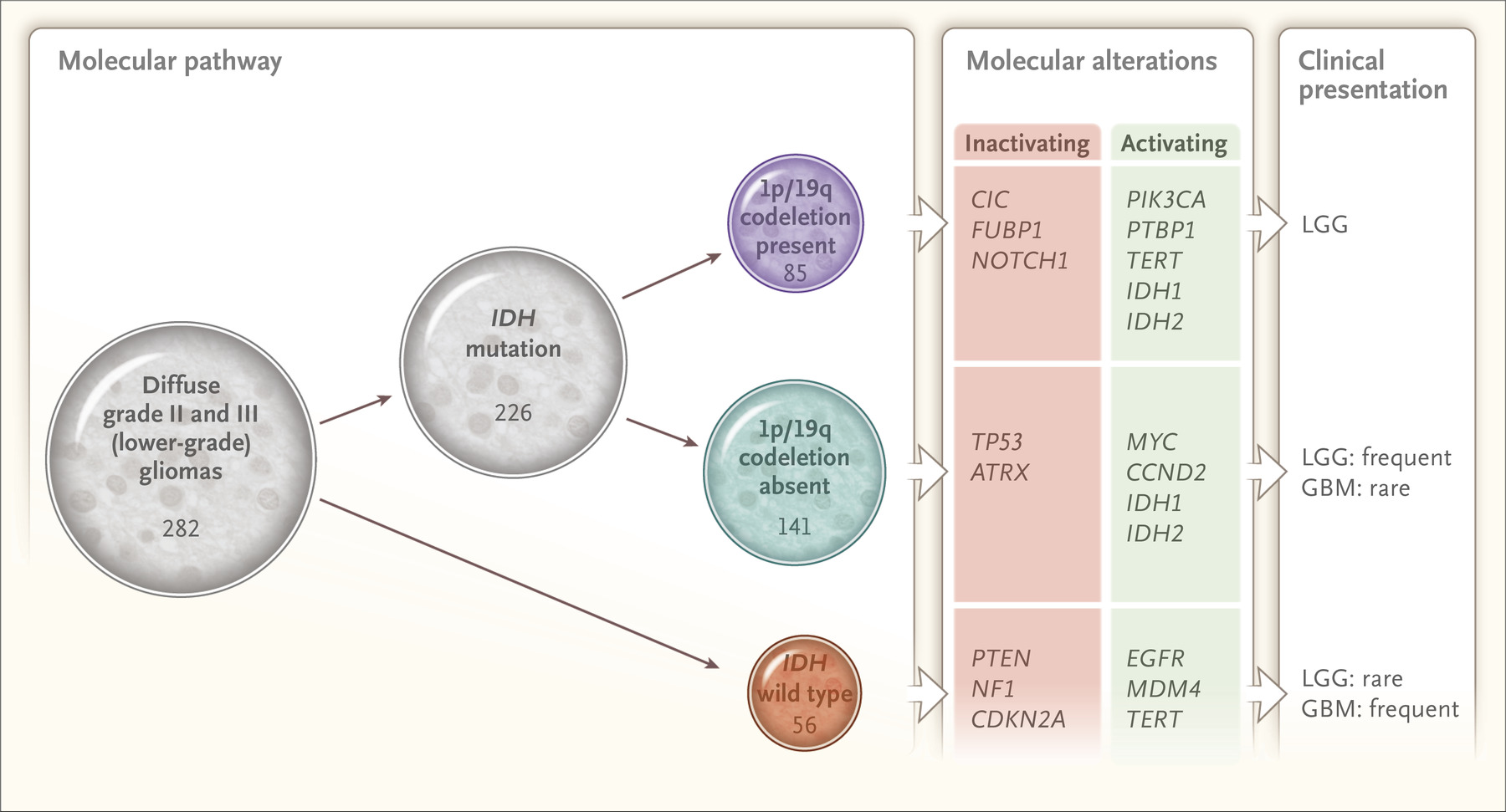
TCGA Low Grade Glioma Paper
The TCGA lower grade glioma paper was published in the New England Journal today with critical contributions from our lab. Roel was cited on NBC news!
TCGA Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015. PMID: 26061751, Full text article
Tumor evolution in glioblastoma
Hoon and Siyuan are co-first authors of our Genome Research publication on tumor evolution in glioblastoma. Congratulations!
Kim H, Zheng S, …, Verhaak RG. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015. PMID: 25650244 Full text article